
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000245407 | SLC22A5-201 | 3237 | XM_011543590 | ENSP00000245407 | 557 (aa) | XP_011541892 | O76082 |
| ENST00000461013 | SLC22A5-207 | 10435 | - | - | - (aa) | - | - |
| ENST00000447841 | SLC22A5-205 | 998 | - | ENSP00000389284 | 37 (aa) | - | H7BZF0 |
| ENST00000435065 | SLC22A5-203 | 1746 | - | ENSP00000402760 | 581 (aa) | - | O76082 |
| ENST00000479605 | SLC22A5-209 | 561 | - | - | - (aa) | - | - |
| ENST00000415928 | SLC22A5-202 | 720 | - | ENSP00000388838 | 240 (aa) | - | H7BZC0 |
| ENST00000448810 | SLC22A5-206 | 852 | - | ENSP00000401860 | 136 (aa) | - | H7C1R8 |
| ENST00000437841 | SLC22A5-204 | 850 | - | ENSP00000400553 | 141 (aa) | - | F8WCC9 |
| ENST00000475308 | SLC22A5-208 | 3693 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa05231 | Choline metabolism in cancer | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000197375 | Eosinophils | 6.554e-005 | - |
| ENSG00000197375 | Fibrinogen | 1.0370000E-063 | 26561523 |
| ENSG00000197375 | Immunoglobulins | 3E-6 | 23382691 |
| ENSG00000197375 | Fatty Acids, Omega-6 | 4E-6 | 24823311 |
| ENSG00000197375 | Crohn Disease | 3E-6 | 17554300 |
| ENSG00000197375 | Metabolism | 4E-29 | 24816252 |
| ENSG00000197375 | Immunoglobulins | 2E-6 | 23382691 |
| ENSG00000197375 | acylcarnitine | 1E-17 | 26068415 |
| ENSG00000197375 | Metabolism | 1E-17 | 26068415 |
| ENSG00000197375 | Asthma | 2E-7 | 20860503 |
| ENSG00000197375 | Metabolism | 3E-12 | 24816252 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000197375 | rs7731390 | 5 | 132386057 | C | IgG glycosylation | 23382691 | [0.22-0.53] unit increase | 0.3771 | EFO_0005193 |
| ENSG00000197375 | rs7731390 | 5 | 132386057 | C | IgG glycosylation | 23382691 | [0.23-0.54] unit increase | 0.3846 | EFO_0005193 |
| ENSG00000197375 | rs274557 | 5 | 132385512 | T | Plasma omega-6 polyunsaturated fatty acid levels (arachidonic acid) | 24823311 | [NR] unit increase | 0.14 | EFO_0005680 |
| ENSG00000197375 | rs274567 | 5 | 132378717 | T | Blood metabolite levels | 24816252 | [0.012-0.024] unit increase | 0.018 | EFO_0005664 |
| ENSG00000197375 | rs2073643 | 5 | 132387596 | T | Mosquito bite size | 28199695 | [0.038-0.059] unit increase | 0.0483183 | EFO_0008378 |
| ENSG00000197375 | rs2073643 | 5 | 132387596 | T | Asthma | 20860503 | [1.06-1.15] | 1.11 | EFO_0000270 |
| ENSG00000197375 | rs11746555 | 5 | 132391341 | ? | Autoimmune traits | 30595370 | EFO_0005140 | ||
| ENSG00000197375 | rs2631367 | 5 | 132369766 | ? | White blood cell count | 30595370 | EFO_0004308 | ||
| ENSG00000197375 | rs10060615 | 5 | 132373185 | C | Diastolic blood pressure | 30578418 | [0.098-0.198] mmHg increase | 0.1483 | EFO_0006336 |
| ENSG00000197375 | rs274555 | 5 | 132387259 | ? | Fat-free mass | 30593698 | [0.094-0.19] unit decrease | 0.14208 | EFO_0004995 |
| ENSG00000197375 | rs274555 | 5 | 132387259 | ? | Fat-free mass | 30593698 | [0.15-0.29] unit decrease | 0.22148 | EFO_0004995 |
| ENSG00000197375 | rs274555 | 5 | 132387259 | ? | Fat-free mass | 30593698 | [0.14-0.23] unit decrease | 0.18633 | EFO_0004995 |
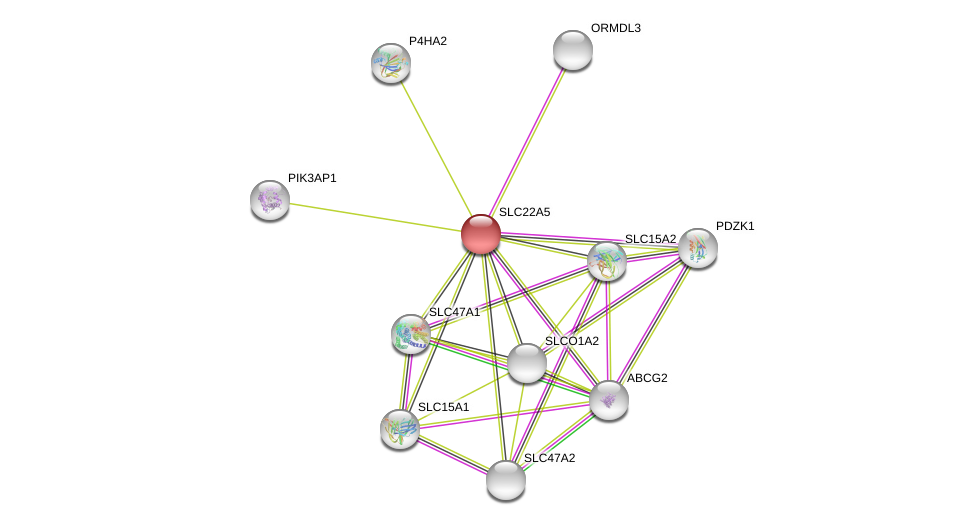
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005515 | protein binding | 15523054. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005886 | plasma membrane | 9618255. | IC | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0006814 | sodium ion transport | - | IEA | Process |
| GO:0006855 | drug transmembrane transport | - | IEA | Process |
| GO:0015226 | carnitine transmembrane transporter activity | 9685390. | EXP | Function |
| GO:0015226 | carnitine transmembrane transporter activity | 9685390. | IDA | Function |
| GO:0015226 | carnitine transmembrane transporter activity | 9916797. | IMP | Function |
| GO:0015238 | drug transmembrane transporter activity | 15238359. | IC | Function |
| GO:0015293 | symporter activity | - | IEA | Function |
| GO:0015651 | quaternary ammonium group transmembrane transporter activity | 9618255. | IDA | Function |
| GO:0015697 | quaternary ammonium group transport | 9618255. | IDA | Process |
| GO:0015879 | carnitine transport | 9685390. | IDA | Process |
| GO:0015879 | carnitine transport | 9916797. | IMP | Process |
| GO:0015893 | drug transport | 15238359. | IC | Process |
| GO:0016021 | integral component of membrane | - | IEA | Component |
| GO:0016324 | apical plasma membrane | 17274673.18641280. | IDA | Component |
| GO:0030165 | PDZ domain binding | 15523054. | IPI | Function |
| GO:0031526 | brush border membrane | 15238359. | IDA | Component |
| GO:0031526 | brush border membrane | 15523054. | ISS | Component |
| GO:0052106 | quorum sensing involved in interaction with host | 18005709. | IMP | Process |
| GO:0060731 | positive regulation of intestinal epithelial structure maintenance | 18005709. | IMP | Process |
| GO:0070062 | extracellular exosome | 19056867. | HDA | Component |
| GO:0070715 | sodium-dependent organic cation transport | 9685390.15238359. | IDA | Process |
| GO:1902603 | carnitine transmembrane transport | - | TAS | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CHOL | |||
| COAD | |||
| KIRP | |||
| LAML | |||
| LUAD | |||
| LUSC | |||
| MESO | |||
| OV | |||
| PAAD | |||
| PRAD | |||
| READ | |||
| STAD | |||
| THCA | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| MESO | ||
| ACC | ||
| PRAD | ||
| KIRP | ||
| PAAD | ||
| PCPG | ||
| BLCA | ||
| READ | ||
| LAML | ||
| UCEC | ||
| GBM | ||
| LIHC |