
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 29554310 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000489328 | GIGYF2-242 | 1116 | - | - | - (aa) | - | - |
| ENST00000409547 | GIGYF2-205 | 5978 | - | ENSP00000386537 | 1299 (aa) | - | Q6Y7W6 |
| ENST00000424038 | GIGYF2-210 | 626 | - | ENSP00000399064 | 92 (aa) | - | F8WCD5 |
| ENST00000428883 | GIGYF2-216 | 544 | - | ENSP00000402712 | 115 (aa) | - | C9JRZ2 |
| ENST00000490229 | GIGYF2-243 | 863 | - | - | - (aa) | - | - |
| ENST00000409196 | GIGYF2-202 | 5747 | - | ENSP00000387070 | 1293 (aa) | - | Q6Y7W6 |
| ENST00000421433 | GIGYF2-207 | 564 | - | ENSP00000402053 | 93 (aa) | - | C9JPV7 |
| ENST00000474312 | GIGYF2-231 | 4687 | - | - | - (aa) | - | - |
| ENST00000490612 | GIGYF2-244 | 567 | - | - | - (aa) | - | - |
| ENST00000426102 | GIGYF2-213 | 561 | - | ENSP00000415037 | 149 (aa) | - | C9JH18 |
| ENST00000473170 | GIGYF2-230 | 550 | - | - | - (aa) | - | - |
| ENST00000421778 | GIGYF2-208 | 564 | - | ENSP00000390325 | 90 (aa) | - | C9JH78 |
| ENST00000427233 | GIGYF2-214 | 530 | - | ENSP00000396590 | 13 (aa) | - | C9JC65 |
| ENST00000409480 | GIGYF2-204 | 5976 | - | ENSP00000386765 | 1321 (aa) | - | I1E4Y6 |
| ENST00000471011 | GIGYF2-229 | 539 | - | - | - (aa) | - | - |
| ENST00000456491 | GIGYF2-223 | 466 | - | ENSP00000413612 | 68 (aa) | - | C9JW88 |
| ENST00000373563 | GIGYF2-201 | 5847 | - | ENSP00000362664 | 1299 (aa) | - | Q6Y7W6 |
| ENST00000484409 | GIGYF2-239 | 367 | - | - | - (aa) | - | - |
| ENST00000469843 | GIGYF2-228 | 550 | - | - | - (aa) | - | - |
| ENST00000488734 | GIGYF2-241 | 796 | - | - | - (aa) | - | - |
| ENST00000475530 | GIGYF2-234 | 287 | - | - | - (aa) | - | - |
| ENST00000423659 | GIGYF2-209 | 2329 | - | ENSP00000404195 | 720 (aa) | - | C9JHW1 |
| ENST00000482952 | GIGYF2-237 | 618 | - | - | - (aa) | - | - |
| ENST00000629305 | GIGYF2-246 | 7878 | - | ENSP00000487548 | 1321 (aa) | - | I1E4Y6 |
| ENST00000427649 | GIGYF2-215 | 677 | - | ENSP00000398055 | 143 (aa) | - | C9J7G1 |
| ENST00000436349 | GIGYF2-219 | 547 | - | ENSP00000400076 | 62 (aa) | - | C9J1A6 |
| ENST00000464805 | GIGYF2-227 | 1196 | NR_103494 | - | - (aa) | - | - |
| ENST00000482666 | GIGYF2-236 | 1261 | NR_103493 | - | - (aa) | - | - |
| ENST00000455139 | GIGYF2-222 | 556 | - | ENSP00000395299 | 62 (aa) | - | A0A1U8QZH8 |
| ENST00000475359 | GIGYF2-233 | 825 | NR_103495 | - | - (aa) | - | - |
| ENST00000424414 | GIGYF2-211 | 583 | - | ENSP00000401261 | 71 (aa) | - | C9JZC0 |
| ENST00000440945 | GIGYF2-220 | 2658 | - | ENSP00000410297 | 836 (aa) | - | E7ESB6 |
| ENST00000458528 | GIGYF2-224 | 566 | - | ENSP00000389322 | 59 (aa) | - | F8WBF1 |
| ENST00000409451 | GIGYF2-203 | 5937 | - | ENSP00000387170 | 1320 (aa) | - | Q6Y7W6 |
| ENST00000483164 | GIGYF2-238 | 577 | - | - | - (aa) | - | - |
| ENST00000430720 | GIGYF2-218 | 561 | - | ENSP00000396958 | 112 (aa) | - | C9JHT0 |
| ENST00000425040 | GIGYF2-212 | 588 | - | ENSP00000407161 | 99 (aa) | - | C9JXQ0 |
| ENST00000445650 | GIGYF2-221 | 588 | - | ENSP00000392218 | 119 (aa) | - | C9IYH5 |
| ENST00000492910 | GIGYF2-245 | 685 | - | - | - (aa) | - | - |
| ENST00000464402 | GIGYF2-226 | 529 | - | - | - (aa) | - | - |
| ENST00000463554 | GIGYF2-225 | 1085 | - | - | - (aa) | - | - |
| ENST00000476995 | GIGYF2-235 | 766 | - | - | - (aa) | - | - |
| ENST00000474465 | GIGYF2-232 | 625 | - | - | - (aa) | - | - |
| ENST00000410033 | GIGYF2-206 | 957 | - | ENSP00000387276 | 176 (aa) | - | B8ZZD2 |
| ENST00000488629 | GIGYF2-240 | 562 | - | - | - (aa) | - | - |
| ENST00000429187 | GIGYF2-217 | 606 | - | ENSP00000392991 | 106 (aa) | - | C9J0V6 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000204120 | rs6704768 | 2 | 232727791 | ? | Autism spectrum disorder or schizophrenia | 28540026 | [1.04-1.09] | 1.0638298 | EFO_0000692|EFO_0003756 |
| ENSG00000204120 | rs6704768 | 2 | 232727791 | G | Schizophrenia | 25056061 | [1.05-1.1] | 1.0752687 | EFO_0000692 |
| ENSG00000204120 | rs1801251 | 2 | 232768750 | A | Coronary artery disease | 29212778 | [0.029-0.055] unit increase | 0.0416 | EFO_0000378 |
| ENSG00000204120 | rs6704768 | 2 | 232727791 | A | Educational attainment (MTAG) | 30038396 | [0.011-0.016] unit decrease | 0.0134 | EFO_0004784 |
| ENSG00000204120 | rs6704768 | 2 | 232727791 | A | Self-reported math ability (MTAG) | 30038396 | [0.016-0.023] unit decrease | 0.0193 | EFO_0004875 |
| ENSG00000204120 | rs6704768 | 2 | 232727791 | A | Highest math class taken (MTAG) | 30038396 | [0.018-0.024] unit decrease | 0.021 | EFO_0004875 |
| ENSG00000204120 | rs12694917 | 2 | 232842901 | T | Cognitive performance (MTAG) | 30038396 | [0.012-0.022] unit increase | 0.017 | EFO_0008354 |
| ENSG00000204120 | rs4144797 | 2 | 232697487 | ? | Schizophrenia | 29483656 | [1.07-1.1] | 1.085 | EFO_0000692 |
| ENSG00000204120 | rs6704768 | 2 | 232727791 | A | Educational attainment (years of education) | 30038396 | [0.0089-0.0143] unit decrease | 0.0116 | EFO_0004784 |
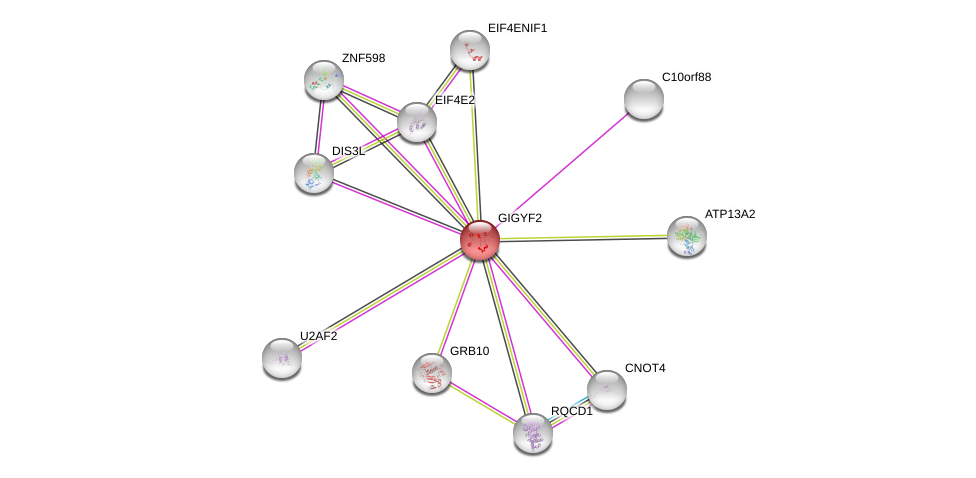
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000204120 | GIGYF2 | 100 | 100.000 | ENSMUSG00000048000 | Gigyf2 | 100 | 96.429 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0005515 | protein binding | 20670374.20696395.20878056.27157137. | IPI | Function |
| GO:0005768 | endosome | 20670374. | IDA | Component |
| GO:0005783 | endoplasmic reticulum | 20696395. | IDA | Component |
| GO:0005794 | Golgi apparatus | 20696395. | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0007631 | feeding behavior | - | IEA | Process |
| GO:0008344 | adult locomotory behavior | - | IEA | Process |
| GO:0009791 | post-embryonic development | - | IEA | Process |
| GO:0010494 | cytoplasmic stress granule | 20696395. | IDA | Component |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016021 | integral component of membrane | - | IEA | Component |
| GO:0016441 | posttranscriptional gene silencing | 27157137. | IDA | Process |
| GO:0017148 | negative regulation of translation | 22751931. | IMP | Process |
| GO:0021522 | spinal cord motor neuron differentiation | - | IEA | Process |
| GO:0031571 | mitotic G1 DNA damage checkpoint | - | IEA | Process |
| GO:0032991 | protein-containing complex | 20878056. | IDA | Component |
| GO:0035264 | multicellular organism growth | - | IEA | Process |
| GO:0043204 | perikaryon | 20670374. | IDA | Component |
| GO:0044267 | cellular protein metabolic process | - | IEA | Process |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0048009 | insulin-like growth factor receptor signaling pathway | 20670374. | IMP | Process |
| GO:0048873 | homeostasis of number of cells within a tissue | - | IEA | Process |
| GO:0050881 | musculoskeletal movement | - | IEA | Process |
| GO:0050885 | neuromuscular process controlling balance | - | IEA | Process |
| GO:0061157 | mRNA destabilization | 27157137. | IDA | Process |
| GO:0070064 | proline-rich region binding | 20696395. | IDA | Function |
| GO:1990635 | proximal dendrite | 20670374. | IDA | Component |