
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000377548 | RRM_5 | PF13893.6 | 2.3e-09 | 1 | 2 |
| ENSP00000377548 | RRM_5 | PF13893.6 | 2.3e-09 | 2 | 2 |
| ENSP00000311747 | RRM_5 | PF13893.6 | 1.8e-07 | 1 | 2 |
| ENSP00000311747 | RRM_5 | PF13893.6 | 1.8e-07 | 2 | 2 |
| ENSP00000377548 | RRM_1 | PF00076.22 | 5.2e-30 | 1 | 2 |
| ENSP00000377548 | RRM_1 | PF00076.22 | 5.2e-30 | 2 | 2 |
| ENSP00000311747 | RRM_1 | PF00076.22 | 4.9e-28 | 1 | 2 |
| ENSP00000311747 | RRM_1 | PF00076.22 | 4.9e-28 | 2 | 2 |
| ENSP00000414650 | RRM_1 | PF00076.22 | 8e-19 | 1 | 2 |
| ENSP00000414650 | RRM_1 | PF00076.22 | 8e-19 | 2 | 2 |
| ENSP00000386995 | RRM_1 | PF00076.22 | 1e-18 | 1 | 2 |
| ENSP00000386995 | RRM_1 | PF00076.22 | 1e-18 | 2 | 2 |
| ENSP00000386518 | RRM_1 | PF00076.22 | 3.6e-18 | 1 | 2 |
| ENSP00000386518 | RRM_1 | PF00076.22 | 3.6e-18 | 2 | 2 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000460762 | RBM14-206 | 459 | - | - | - (aa) | - | - |
| ENST00000443702 | RBM14-205 | 732 | - | ENSP00000414650 | 129 (aa) | - | F8WDX3 |
| ENST00000461478 | RBM14-207 | 693 | - | - | - (aa) | - | - |
| ENST00000310137 | RBM14-201 | 2828 | - | ENSP00000311747 | 669 (aa) | - | Q96PK6 |
| ENST00000409372 | RBM14-203 | 876 | - | ENSP00000386518 | 159 (aa) | - | B8ZZ74 |
| ENST00000409738 | RBM14-204 | 372 | - | ENSP00000386995 | 119 (aa) | - | Q96PK6 |
| ENST00000496694 | RBM14-208 | 562 | - | - | - (aa) | - | - |
| ENST00000512283 | RBM14-209 | 567 | - | - | - (aa) | - | - |
| ENST00000393979 | RBM14-202 | 1426 | - | ENSP00000377548 | 156 (aa) | - | Q96PK6 |
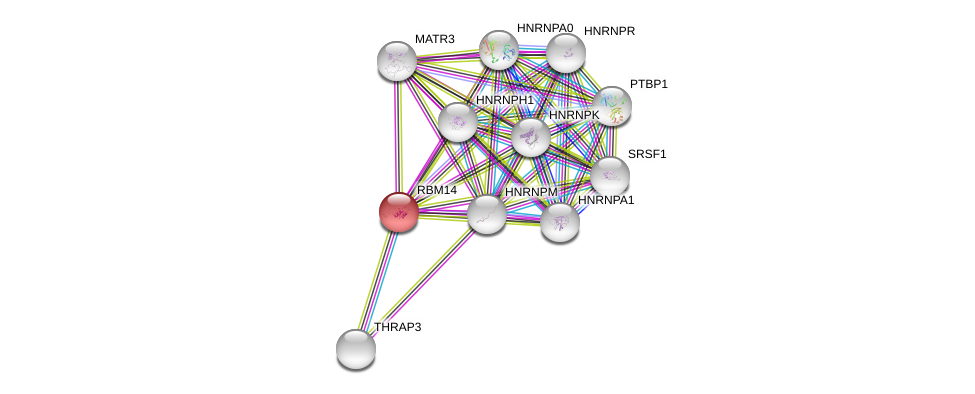
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000239306 | RBM14 | 98 | 31.061 | ENSG00000135486 | HNRNPA1 | 57 | 32.090 |
| ENSG00000239306 | RBM14 | 94 | 48.529 | ENSG00000173914 | RBM4B | 98 | 48.529 |
| ENSG00000239306 | RBM14 | 94 | 48.529 | ENSG00000173933 | RBM4 | 97 | 48.529 |
| ENSG00000239306 | RBM14 | 53 | 36.264 | ENSG00000213516 | RBMXL1 | 54 | 36.264 |
| ENSG00000239306 | RBM14 | 53 | 36.264 | ENSG00000147274 | RBMX | 92 | 33.766 |
| ENSG00000239306 | RBM14 | 52 | 30.488 | ENSG00000125970 | RALY | 90 | 30.864 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000239306 | RBM14 | 96 | 61.842 | ENSACAG00000012696 | RBM14 | 99 | 46.011 | Anolis_carolinensis |
| ENSG00000239306 | RBM14 | 95 | 39.437 | ENSACAG00000028084 | - | 86 | 40.141 | Anolis_carolinensis |
| ENSG00000239306 | RBM14 | 100 | 99.851 | ENSANAG00000035377 | RBM14 | 100 | 99.851 | Aotus_nancymaae |
| ENSG00000239306 | RBM14 | 100 | 99.253 | ENSBTAG00000001225 | RBM14 | 100 | 99.253 | Bos_taurus |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSCJAG00000001294 | RBM14 | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000239306 | RBM14 | 100 | 99.701 | ENSCAFG00000012405 | RBM14 | 100 | 99.701 | Canis_familiaris |
| ENSG00000239306 | RBM14 | 100 | 99.701 | ENSCAFG00020015516 | RBM14 | 100 | 99.701 | Canis_lupus_dingo |
| ENSG00000239306 | RBM14 | 100 | 99.253 | ENSCHIG00000020952 | RBM14 | 100 | 99.253 | Capra_hircus |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSTSYG00000000450 | RBM14 | 100 | 100.000 | Carlito_syrichta |
| ENSG00000239306 | RBM14 | 85 | 96.041 | ENSCAPG00000012991 | RBM14 | 96 | 96.041 | Cavia_aperea |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSCPOG00000008019 | RBM14 | 100 | 100.000 | Cavia_porcellus |
| ENSG00000239306 | RBM14 | 100 | 97.436 | ENSCCAG00000034251 | - | 100 | 97.436 | Cebus_capucinus |
| ENSG00000239306 | RBM14 | 100 | 99.552 | ENSCCAG00000019303 | RBM14 | 100 | 99.552 | Cebus_capucinus |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSCATG00000036313 | RBM14 | 100 | 100.000 | Cercocebus_atys |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSCLAG00000015277 | RBM14 | 100 | 100.000 | Chinchilla_lanigera |
| ENSG00000239306 | RBM14 | 99 | 96.610 | ENSCSAG00000008356 | - | 77 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000239306 | RBM14 | 95 | 68.456 | ENSCPBG00000023808 | RBM14 | 99 | 53.163 | Chrysemys_picta_bellii |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSCANG00000028611 | RBM14 | 100 | 100.000 | Colobus_angolensis_palliatus |
| ENSG00000239306 | RBM14 | 100 | 99.160 | ENSCGRG00001024921 | Rbm14 | 100 | 99.160 | Cricetulus_griseus_chok1gshd |
| ENSG00000239306 | RBM14 | 100 | 99.160 | ENSCGRG00000010784 | Rbm14 | 100 | 99.160 | Cricetulus_griseus_crigri |
| ENSG00000239306 | RBM14 | 100 | 98.954 | ENSDNOG00000043225 | RBM14 | 100 | 98.954 | Dasypus_novemcinctus |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSDORG00000002474 | Rbm14 | 100 | 100.000 | Dipodomys_ordii |
| ENSG00000239306 | RBM14 | 99 | 42.857 | ENSEBUG00000008661 | - | 61 | 43.226 | Eptatretus_burgeri |
| ENSG00000239306 | RBM14 | 100 | 99.701 | ENSEASG00005005062 | RBM14 | 100 | 99.701 | Equus_asinus_asinus |
| ENSG00000239306 | RBM14 | 100 | 99.701 | ENSECAG00000022859 | RBM14 | 100 | 99.701 | Equus_caballus |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSFCAG00000007392 | RBM14 | 100 | 100.000 | Felis_catus |
| ENSG00000239306 | RBM14 | 95 | 65.541 | ENSFALG00000002071 | RBM14 | 100 | 41.134 | Ficedula_albicollis |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSFDAG00000002787 | RBM14 | 100 | 100.000 | Fukomys_damarensis |
| ENSG00000239306 | RBM14 | 99 | 53.165 | ENSGMOG00000009633 | rbm14b | 69 | 53.165 | Gadus_morhua |
| ENSG00000239306 | RBM14 | 100 | 65.854 | ENSGALG00000035260 | RBM14 | 96 | 53.226 | Gallus_gallus |
| ENSG00000239306 | RBM14 | 99 | 54.140 | ENSGACG00000019028 | rbm14b | 69 | 54.140 | Gasterosteus_aculeatus |
| ENSG00000239306 | RBM14 | 87 | 51.592 | ENSGAGG00000008555 | - | 99 | 50.929 | Gopherus_agassizii |
| ENSG00000239306 | RBM14 | 99 | 62.185 | ENSGAGG00000001366 | - | 84 | 62.185 | Gopherus_agassizii |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSGGOG00000014455 | RBM14 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000239306 | RBM14 | 100 | 99.253 | ENSHGLG00000012589 | RBM14 | 100 | 99.253 | Heterocephalus_glaber_female |
| ENSG00000239306 | RBM14 | 100 | 99.103 | ENSHGLG00100011836 | RBM14 | 100 | 99.103 | Heterocephalus_glaber_male |
| ENSG00000239306 | RBM14 | 99 | 53.797 | ENSHCOG00000012571 | rbm14b | 97 | 57.018 | Hippocampus_comes |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSSTOG00000009796 | RBM14 | 100 | 100.000 | Ictidomys_tridecemlineatus |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSJJAG00000020814 | Rbm14 | 100 | 100.000 | Jaculus_jaculus |
| ENSG00000239306 | RBM14 | 99 | 53.548 | ENSLACG00000017750 | - | 54 | 53.947 | Latimeria_chalumnae |
| ENSG00000239306 | RBM14 | 84 | 92.665 | ENSLAFG00000009086 | RBM14 | 100 | 92.665 | Loxodonta_africana |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSMFAG00000003827 | RBM4 | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSMMUG00000020363 | RBM14 | 100 | 100.000 | Macaca_mulatta |
| ENSG00000239306 | RBM14 | 100 | 99.851 | ENSMNEG00000014872 | RBM14 | 100 | 99.851 | Macaca_nemestrina |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSMLEG00000024601 | RBM14 | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000239306 | RBM14 | 97 | 63.248 | ENSMGAG00000016129 | - | 97 | 63.248 | Meleagris_gallopavo |
| ENSG00000239306 | RBM14 | 100 | 99.160 | ENSMAUG00000017509 | Rbm14 | 100 | 99.160 | Mesocricetus_auratus |
| ENSG00000239306 | RBM14 | 100 | 99.552 | ENSMICG00000031929 | RBM14 | 100 | 99.552 | Microcebus_murinus |
| ENSG00000239306 | RBM14 | 100 | 99.160 | ENSMOCG00000002491 | Rbm14 | 100 | 99.160 | Microtus_ochrogaster |
| ENSG00000239306 | RBM14 | 90 | 94.366 | ENSMODG00000007884 | RBM14 | 100 | 84.628 | Monodelphis_domestica |
| ENSG00000239306 | RBM14 | 100 | 50.610 | ENSMALG00000019079 | - | 96 | 50.610 | Monopterus_albus |
| ENSG00000239306 | RBM14 | 100 | 99.160 | MGP_CAROLIEiJ_G0022517 | Rbm14 | 100 | 99.160 | Mus_caroli |
| ENSG00000239306 | RBM14 | 100 | 99.160 | ENSMUSG00000006456 | Rbm14 | 100 | 99.160 | Mus_musculus |
| ENSG00000239306 | RBM14 | 100 | 99.160 | MGP_PahariEiJ_G0014014 | Rbm14 | 100 | 99.160 | Mus_pahari |
| ENSG00000239306 | RBM14 | 100 | 99.160 | MGP_SPRETEiJ_G0023431 | Rbm14 | 100 | 99.160 | Mus_spretus |
| ENSG00000239306 | RBM14 | 100 | 99.701 | ENSMPUG00000003440 | RBM14 | 100 | 99.701 | Mustela_putorius_furo |
| ENSG00000239306 | RBM14 | 100 | 94.359 | ENSMLUG00000024395 | - | 100 | 74.141 | Myotis_lucifugus |
| ENSG00000239306 | RBM14 | 100 | 98.505 | ENSMLUG00000015545 | - | 100 | 98.505 | Myotis_lucifugus |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSNGAG00000019304 | Rbm14 | 100 | 100.000 | Nannospalax_galili |
| ENSG00000239306 | RBM14 | 100 | 99.676 | ENSNLEG00000006197 | RBM14 | 95 | 99.676 | Nomascus_leucogenys |
| ENSG00000239306 | RBM14 | 85 | 85.140 | ENSMEUG00000005742 | RBM14 | 100 | 85.664 | Notamacropus_eugenii |
| ENSG00000239306 | RBM14 | 100 | 98.057 | ENSOPRG00000009139 | RBM14 | 100 | 98.057 | Ochotona_princeps |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSODEG00000017717 | RBM14 | 100 | 100.000 | Octodon_degus |
| ENSG00000239306 | RBM14 | 97 | 90.517 | ENSOANG00000001240 | - | 97 | 90.517 | Ornithorhynchus_anatinus |
| ENSG00000239306 | RBM14 | 100 | 99.103 | ENSOCUG00000022838 | RBM14 | 100 | 99.103 | Oryctolagus_cuniculus |
| ENSG00000239306 | RBM14 | 100 | 47.899 | ENSOMEG00000006508 | - | 93 | 48.980 | Oryzias_melastigma |
| ENSG00000239306 | RBM14 | 100 | 99.253 | ENSOGAG00000032829 | RBM14 | 100 | 99.253 | Otolemur_garnettii |
| ENSG00000239306 | RBM14 | 100 | 93.890 | ENSOARG00000007565 | RBM14 | 100 | 93.890 | Ovis_aries |
| ENSG00000239306 | RBM14 | 100 | 99.851 | ENSPPAG00000033403 | RBM14 | 100 | 99.851 | Pan_paniscus |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSPPRG00000003716 | RBM14 | 100 | 100.000 | Panthera_pardus |
| ENSG00000239306 | RBM14 | 86 | 99.476 | ENSPTIG00000016775 | RBM14 | 86 | 99.476 | Panthera_tigris_altaica |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSPTRG00000003934 | RBM14 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSPANG00000004924 | RBM14 | 100 | 100.000 | Papio_anubis |
| ENSG00000239306 | RBM14 | 81 | 80.000 | ENSPSIG00000003436 | RBM14 | 99 | 53.106 | Pelodiscus_sinensis |
| ENSG00000239306 | RBM14 | 100 | 47.899 | ENSPMGG00000018869 | - | 91 | 48.966 | Periophthalmus_magnuspinnatus |
| ENSG00000239306 | RBM14 | 96 | 94.000 | ENSPCIG00000008541 | RBM14 | 92 | 94.000 | Phascolarctos_cinereus |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSPPYG00000003015 | RBM14 | 100 | 100.000 | Pongo_abelii |
| ENSG00000239306 | RBM14 | 96 | 99.333 | ENSPCAG00000003334 | RBM14 | 90 | 98.492 | Procavia_capensis |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSPCOG00000016329 | RBM14 | 100 | 100.000 | Propithecus_coquereli |
| ENSG00000239306 | RBM14 | 96 | 100.000 | ENSPVAG00000010483 | RBM14 | 74 | 97.872 | Pteropus_vampyrus |
| ENSG00000239306 | RBM14 | 100 | 98.655 | ENSRNOG00000045568 | Rbm14 | 100 | 98.655 | Rattus_norvegicus |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSRBIG00000043267 | RBM14 | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSRROG00000044302 | RBM14 | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000239306 | RBM14 | 100 | 100.000 | ENSSBOG00000031468 | RBM14 | 100 | 100.000 | Saimiri_boliviensis_boliviensis |
| ENSG00000239306 | RBM14 | 84 | 80.836 | ENSSHAG00000008478 | RBM14 | 100 | 80.662 | Sarcophilus_harrisii |
| ENSG00000239306 | RBM14 | 94 | 100.000 | ENSSARG00000006530 | RBM14 | 100 | 92.526 | Sorex_araneus |
| ENSG00000239306 | RBM14 | 96 | 74.138 | ENSSPUG00000005905 | RBM14 | 100 | 53.754 | Sphenodon_punctatus |
| ENSG00000239306 | RBM14 | 100 | 99.552 | ENSSSCG00000038008 | RBM14 | 100 | 99.552 | Sus_scrofa |
| ENSG00000239306 | RBM14 | 96 | 96.000 | ENSTBEG00000003641 | RBM14 | 100 | 91.330 | Tupaia_belangeri |
| ENSG00000239306 | RBM14 | 96 | 100.000 | ENSTTRG00000011156 | RBM14 | 89 | 98.881 | Tursiops_truncatus |
| ENSG00000239306 | RBM14 | 96 | 99.333 | ENSUAMG00000014883 | RBM14 | 99 | 98.686 | Ursus_americanus |
| ENSG00000239306 | RBM14 | 80 | 90.467 | ENSUMAG00000011736 | RBM14 | 95 | 90.467 | Ursus_maritimus |
| ENSG00000239306 | RBM14 | 96 | 100.000 | ENSVVUG00000010624 | RBM14 | 100 | 99.672 | Vulpes_vulpes |
| ENSG00000239306 | RBM14 | 96 | 64.430 | ENSXETG00000026650 | rbm14 | 99 | 40.310 | Xenopus_tropicalis |
| ENSG00000239306 | RBM14 | 98 | 45.098 | ENSXETG00000030260 | - | 56 | 45.614 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0002218 | activation of innate immune response | 21873635. | IBA | Process |
| GO:0002218 | activation of innate immune response | 28712728. | IDA | Process |
| GO:0003712 | transcription coregulator activity | 11443112. | NAS | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003723 | RNA binding | 21873635. | IBA | Function |
| GO:0003723 | RNA binding | 11443112. | NAS | Function |
| GO:0005515 | protein binding | 15919756.25385835.26871637. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 25385835. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005667 | transcription factor complex | 21873635. | IBA | Component |
| GO:0005667 | transcription factor complex | 11443112. | IPI | Component |
| GO:0005730 | nucleolus | - | IEA | Component |
| GO:0005737 | cytoplasm | 25385835. | IDA | Component |
| GO:0006260 | DNA replication | 11443112. | NAS | Process |
| GO:0006281 | DNA repair | 11443112. | NAS | Process |
| GO:0006310 | DNA recombination | 11443112. | NAS | Process |
| GO:0009725 | response to hormone | 11443112. | TAS | Process |
| GO:0016575 | histone deacetylation | 11443112. | IPI | Process |
| GO:0016592 | mediator complex | 11443112. | NAS | Component |
| GO:0016607 | nuclear speck | 21873635. | IBA | Component |
| GO:0016607 | nuclear speck | - | IDA | Component |
| GO:0030374 | nuclear receptor transcription coactivator activity | 21873635. | IBA | Function |
| GO:0030374 | nuclear receptor transcription coactivator activity | 11443112. | IPI | Function |
| GO:0030520 | intracellular estrogen receptor signaling pathway | 11443112. | NAS | Process |
| GO:0030674 | protein binding, bridging | 11443112. | NAS | Function |
| GO:0042921 | glucocorticoid receptor signaling pathway | 11443112. | NAS | Process |
| GO:0045087 | innate immune response | - | IEA | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 21873635. | IBA | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 11443112.15919756. | IDA | Process |
| GO:0046600 | negative regulation of centriole replication | 25385835. | IMP | Process |
| GO:0060395 | SMAD protein signal transduction | - | IEA | Process |
| GO:0098534 | centriole assembly | 25385835. | IMP | Process |
| GO:1990904 | ribonucleoprotein complex | 11443112. | TAS | Component |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ESCA | |||
| TGCT |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| MESO | ||
| ACC | ||
| UCS | ||
| HNSC | ||
| SKCM | ||
| PRAD | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| READ | ||
| LAML | ||
| UCEC | ||
| LIHC | ||
| LUAD |