
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000470185 | INT_SG_DDX_CT_C | PF15300.6 | 1.1e-24 | 1 | 1 |
| ENSP00000472303 | INT_SG_DDX_CT_C | PF15300.6 | 1.1e-24 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000594117 | CT45A1-201 | 1087 | XM_011531352 | ENSP00000470185 | 189 (aa) | XP_011529654 | Q5HYN5 |
| ENST00000594565 | CT45A1-202 | 1007 | XM_005278141 | ENSP00000472303 | 189 (aa) | XP_005278198 | Q5HYN5 |
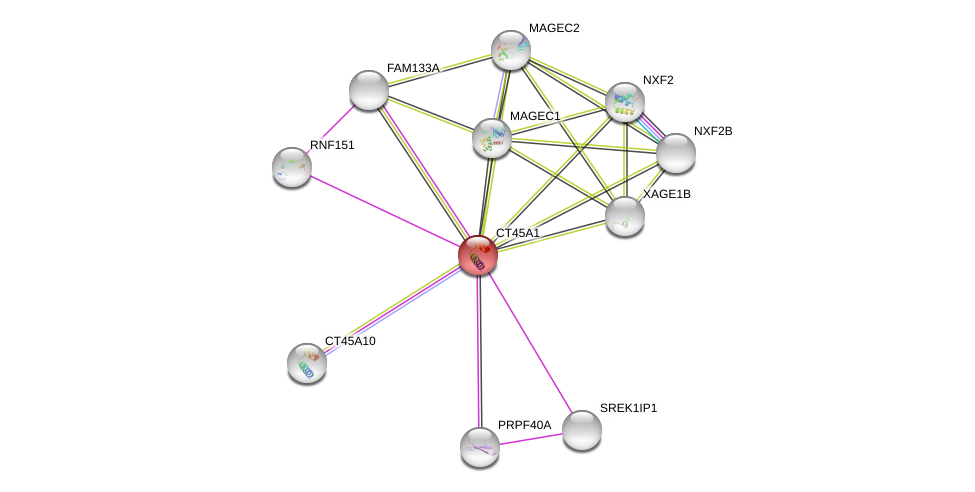
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000268940 | CT45A1 | 69 | 36.765 | ENSAMEG00000005934 | - | 80 | 36.765 | Ailuropoda_melanoleuca |
| ENSG00000268940 | CT45A1 | 98 | 68.817 | ENSANAG00000006559 | - | 98 | 68.817 | Aotus_nancymaae |
| ENSG00000268940 | CT45A1 | 99 | 66.489 | ENSCJAG00000004135 | - | 99 | 66.489 | Callithrix_jacchus |
| ENSG00000268940 | CT45A1 | 92 | 30.857 | ENSCAFG00000014436 | - | 90 | 30.857 | Canis_familiaris |
| ENSG00000268940 | CT45A1 | 87 | 39.535 | ENSCAFG00020000069 | - | 61 | 39.535 | Canis_lupus_dingo |
| ENSG00000268940 | CT45A1 | 85 | 43.529 | ENSCAFG00020024726 | - | 76 | 43.529 | Canis_lupus_dingo |
| ENSG00000268940 | CT45A1 | 100 | 50.259 | ENSTSYG00000003411 | - | 99 | 50.259 | Carlito_syrichta |
| ENSG00000268940 | CT45A1 | 100 | 79.365 | ENSCATG00000026275 | - | 89 | 79.365 | Cercocebus_atys |
| ENSG00000268940 | CT45A1 | 100 | 78.836 | ENSCSAG00000007640 | - | 93 | 78.836 | Chlorocebus_sabaeus |
| ENSG00000268940 | CT45A1 | 100 | 66.138 | ENSCSAG00000015072 | - | 100 | 66.138 | Chlorocebus_sabaeus |
| ENSG00000268940 | CT45A1 | 100 | 77.249 | ENSCANG00000040837 | - | 99 | 77.249 | Colobus_angolensis_palliatus |
| ENSG00000268940 | CT45A1 | 68 | 34.559 | ENSCGRG00001016867 | Gm648 | 60 | 34.559 | Cricetulus_griseus_chok1gshd |
| ENSG00000268940 | CT45A1 | 68 | 34.559 | ENSCGRG00000002325 | Gm648 | 58 | 34.559 | Cricetulus_griseus_crigri |
| ENSG00000268940 | CT45A1 | 100 | 79.365 | ENSMFAG00000035558 | - | 93 | 79.365 | Macaca_fascicularis |
| ENSG00000268940 | CT45A1 | 96 | 77.348 | ENSMFAG00000036609 | - | 100 | 77.348 | Macaca_fascicularis |
| ENSG00000268940 | CT45A1 | 100 | 78.836 | ENSMMUG00000021984 | - | 91 | 78.836 | Macaca_mulatta |
| ENSG00000268940 | CT45A1 | 100 | 52.910 | ENSMMUG00000042170 | - | 100 | 52.910 | Macaca_mulatta |
| ENSG00000268940 | CT45A1 | 100 | 79.894 | ENSMNEG00000028699 | - | 93 | 79.894 | Macaca_nemestrina |
| ENSG00000268940 | CT45A1 | 96 | 77.348 | ENSMNEG00000040510 | - | 100 | 77.348 | Macaca_nemestrina |
| ENSG00000268940 | CT45A1 | 100 | 79.365 | ENSMLEG00000000289 | - | 93 | 79.365 | Mandrillus_leucophaeus |
| ENSG00000268940 | CT45A1 | 69 | 74.615 | ENSMLEG00000042160 | - | 68 | 74.615 | Mandrillus_leucophaeus |
| ENSG00000268940 | CT45A1 | 99 | 32.271 | MGP_CAROLIEiJ_G0033085 | Ints6l | 77 | 49.153 | Mus_caroli |
| ENSG00000268940 | CT45A1 | 71 | 34.965 | MGP_CAROLIEiJ_G0033087 | Gm648 | 64 | 36.111 | Mus_caroli |
| ENSG00000268940 | CT45A1 | 99 | 32.271 | ENSMUSG00000035967 | Ints6l | 76 | 50.847 | Mus_musculus |
| ENSG00000268940 | CT45A1 | 71 | 34.028 | ENSMUSG00000064016 | Gm648 | 64 | 34.028 | Mus_musculus |
| ENSG00000268940 | CT45A1 | 99 | 32.271 | MGP_PahariEiJ_G0031627 | Ints6l | 77 | 50.847 | Mus_pahari |
| ENSG00000268940 | CT45A1 | 71 | 36.111 | MGP_SPRETEiJ_G0034242 | Gm648 | 64 | 36.111 | Mus_spretus |
| ENSG00000268940 | CT45A1 | 99 | 34.400 | MGP_SPRETEiJ_G0034240 | Ints6l | 77 | 50.847 | Mus_spretus |
| ENSG00000268940 | CT45A1 | 100 | 86.772 | ENSNLEG00000002303 | - | 100 | 86.772 | Nomascus_leucogenys |
| ENSG00000268940 | CT45A1 | 100 | 43.005 | ENSOCUG00000027335 | - | 93 | 43.005 | Oryctolagus_cuniculus |
| ENSG00000268940 | CT45A1 | 99 | 37.566 | ENSOARG00000011598 | - | 91 | 37.566 | Ovis_aries |
| ENSG00000268940 | CT45A1 | 100 | 70.899 | ENSPTRG00000028196 | - | 100 | 70.899 | Pan_troglodytes |
| ENSG00000268940 | CT45A1 | 100 | 94.709 | ENSPTRG00000047355 | - | 100 | 94.709 | Pan_troglodytes |
| ENSG00000268940 | CT45A1 | 100 | 94.709 | ENSPTRG00000047577 | CT45A10 | 100 | 94.709 | Pan_troglodytes |
| ENSG00000268940 | CT45A1 | 100 | 79.365 | ENSPANG00000014397 | - | 89 | 79.365 | Papio_anubis |
| ENSG00000268940 | CT45A1 | 100 | 78.307 | ENSPANG00000023647 | - | 99 | 78.307 | Papio_anubis |
| ENSG00000268940 | CT45A1 | 100 | 74.737 | ENSPPYG00000020754 | - | 99 | 74.737 | Pongo_abelii |
| ENSG00000268940 | CT45A1 | 100 | 90.476 | ENSPPYG00000020755 | - | 100 | 90.476 | Pongo_abelii |
| ENSG00000268940 | CT45A1 | 90 | 87.719 | ENSPPYG00000020753 | - | 99 | 87.719 | Pongo_abelii |
| ENSG00000268940 | CT45A1 | 69 | 43.846 | ENSPVAG00000002652 | - | 96 | 43.846 | Pteropus_vampyrus |
| ENSG00000268940 | CT45A1 | 99 | 79.787 | ENSRBIG00000006469 | - | 99 | 79.787 | Rhinopithecus_bieti |
| ENSG00000268940 | CT45A1 | 100 | 78.307 | ENSRROG00000017978 | - | 93 | 78.307 | Rhinopithecus_roxellana |
| ENSG00000268940 | CT45A1 | 99 | 67.553 | ENSSBOG00000024560 | - | 99 | 67.553 | Saimiri_boliviensis_boliviensis |
| ENSG00000268940 | CT45A1 | 96 | 38.021 | ENSUAMG00000005870 | - | 99 | 38.021 | Ursus_americanus |
| ENSG00000268940 | CT45A1 | 99 | 37.563 | ENSUAMG00000003018 | - | 93 | 37.563 | Ursus_americanus |
| ENSG00000268940 | CT45A1 | 89 | 36.872 | ENSUAMG00000021456 | - | 83 | 36.872 | Ursus_americanus |
| ENSG00000268940 | CT45A1 | 51 | 46.392 | ENSVPAG00000011549 | - | 60 | 44.538 | Vicugna_pacos |
| ENSG00000268940 | CT45A1 | 99 | 32.105 | ENSVVUG00000010778 | - | 91 | 32.105 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0032039 | integrator complex | 21873635. | IBA | Component |
| GO:0034472 | snRNA 3'-end processing | 21873635. | IBA | Process |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| HNSC | |||||||
| HNSC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| KIRC | |||
| READ | |||
| TGCT | |||
| THCA | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| UCS | ||
| LGG | ||
| OV |